A study published in Nature Communications reveals the impact of the environment on the genetic adaptation of bacteria. Environments rich in nutrients favor those that have evolved by losing genes involved in metabolic processes such as the self-production of amino acids. Streamlining the genome represents a significant energy saving, giving them an advantage in contexts where these nutrients are readily available.
In the scientific article, researchers from the Universities of Colorado, Aalborg, the Lawrence Berkeley Lab, and the Advanced Studies Center of Blanes (CEAB-CSIC) unveil crucial connections between the environment, survival strategies, and the genetic evolution of bacteria.
To achieve this, the team analyzed information stored in over 26,000 bacterial genomes and 3,800 metagenomes from natural environments such as soils, oceans, lakes, water treatment plants, human and plant microbiotas, as well as foods like sourdough or cheese, using supercomputing. The goal was to correlate genes, survival strategies, and the environment to determine conditions favouring the loss of genes related to amino acid production and the consequent energy saving for survival.
The study reveals that the majority of bacteria—80%, a higher percentage than previously believed—can produce the necessary amino acids for their functioning on their own. The remaining 20% cannot and depend on interaction with other living organisms or with the environment. Similar to humans, this second type of microorganism is obliged to obtain essential compounds through the diet, and they are called “auxotrophs.”
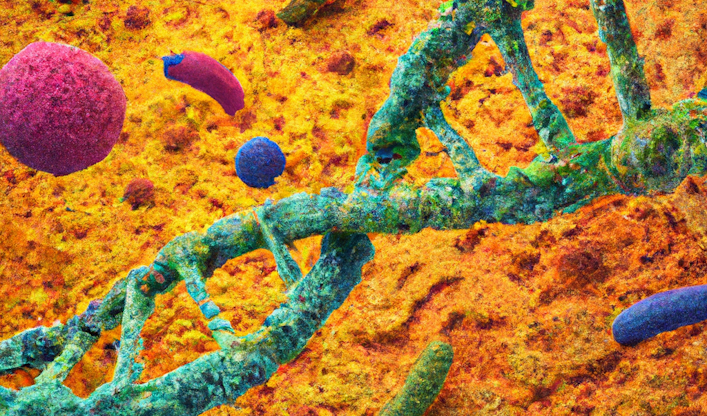
The research highlights that auxotrophic bacteria thrive in environments rich in amino acids, such as human intestines or foods like dairy products. In these nutrient-rich environments, there is a tendency for the evolution towards the loss of genes responsible for amino acid production.
Josep Ramoneda, the lead author of the article, explains the reason simply: “Why make an effort when everything is within reach? Having constant access to a ‘free buffet’ makes self-production of amino acids no longer an advantage. These environments have favored a genome optimization strategy in which bacteria have eliminated unnecessary genes. This has helped them minimize energy costs and thrive. A radical example are mycoplasmas, implicated in numerous diseases, which obtain amino acids from the cells of the humans they parasitize.“
The researchers also found that those bacteria depending on external supply have significantly smaller genomes than those that are self-sufficient, and are lacking genes for other metabolisms like those for sugars or lipids. This suggests that the loss of genes related to amino acid production is just one part of a broader genome rationalization strategy.
The work demonstrates that genomic optimization, due to the significant energy savings it entails, can be a winning strategy in nutrient-rich ecosystems. It also highlights that, in most environments, especially those lacking nutrients, self-sufficiency remains the best option.
Emilio O. Casamayor, a researcher at CEAB-CSIC and co-author of the study, points out that “in aquatic environments, where finding food is more challenging or intermittent, such as in high mountain lakes or the ocean, we have shown that genome rationalization is not the successful strategy. These environments favour bacteria that are self-sufficient.” He adds that “research like this is possible thanks to the combination of massive sequencing and computing techniques, allowing us to tackle more predictive ecological studies in the microbial world on large scales, approaches that were obscured until recently.”
The researchers urge further exploration of the relationship between the environment and the evolutionary adaptation of microbes to advance our understanding of microbial life through increasingly predictive approaches.